Integrated Computational Materials Engineering (ICME)
Particle Characterization with ImageJ
Abstract
This is a tutorial to illustrate the basic capability of imageJ. It's modified from a tutorial on ImageJ's website.
In this tutorial, we'll demonstrate how to use ImageJ to extract particles and determine size distributions of the particles from a digital image.
Tutorial
First, You'll need to download the ImageJ software from here and install it.
1. Open embryos image via Select File → Open Samples → Embryos

2. Draw line over the scale bar and select Analyze → Set Scale, In Set Scale window enter 100 into the 'Known Distance' box and Change the 'Unit of Measurement' box to 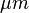 , check 'Global'.
, check 'Global'.
3. Convert the image to grayscale: Image → Type → 8-bit
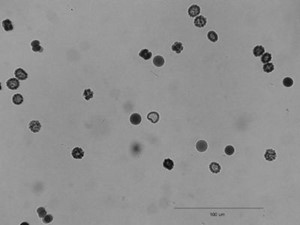
4. Segment the image using the automated routine: Process → Binary → Make Binary
Alternatively you can segment the image manually with Image → Adjust → Threshold and select a threshold range of 0 to 121.
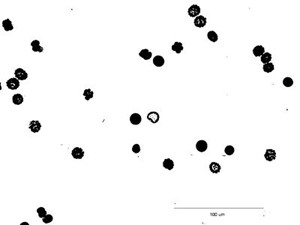
5. Fill the holes in particles: Process → Binary → Fill holes
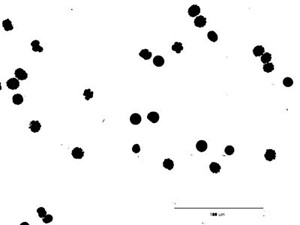
6. Surround the scale bar with the rectangular selection tool and clear the contents (Edit → Clear).
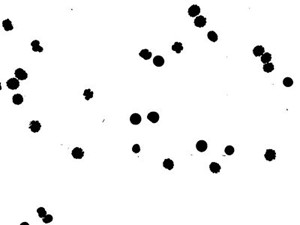
7. Notice in the binary image, there are several particles connected together. Use eraser tool to separate the connected particle manually.
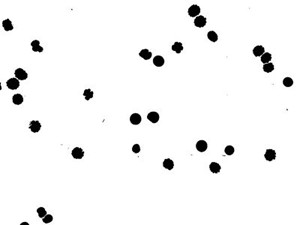
8. Analyze particles: Analyze → Analyze Particles
Enter 20 as the minimum particle size, toggle 'Show Outlines', check 'exclude on edges', ‘Display Results’, ‘Summarize’ and ‘Record Stats’ and click 'OK'. 28 embryos are counted, numbered. The data window lists the area (in 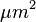 ) for each embryo. These data could be copied to a spreadsheet. A summary of the particle count is also shown in another data window.
) for each embryo. These data could be copied to a spreadsheet. A summary of the particle count is also shown in another data window.

